SignedRangeGenotype Class Reference
SignedRangeGenotype class. More...

Public Member Functions | |
| SignedRangeGenotype (unsigned int numGenes=0, double max=+1, unsigned int bitPrecision=8) | |
| Construct a un-initialized SignedRangeGenotype | |
| SignedRangeGenotype (const SignedRangeGenotype &genotype) | |
| Copy-Constructor (generate a clone of the genoma) | |
| virtual | ~SignedRangeGenotype () |
| Destructor. | |
| virtual double | at (unsigned int i) |
| Return the i-th genes represented. | |
| virtual SignedRangeGenotype * | clone () const |
| Create an exact copy of this Genotype. | |
| virtual void | configure (ConfigurationParameters ¶ms, QString prefix) |
| Configure the object using the ConfigurationParameters specified. | |
| virtual SignedRangeGenotype & | operator= (const Genotype &genotype) |
| Assign operator (performs same operation of Copy-constructor) | |
| virtual void | save (ConfigurationParameters ¶ms, QString prefix) |
| Save the actual status of parameters into the ConfigurationParameters object passed. | |
| virtual void | set (unsigned int i, double value) |
| Set the i-th genes. | |
Static Public Member Functions | |
| static void | describe (QString type) |
| Add to Factory::typeDescriptions() the descriptions of all parameters and subgroups. | |
Detailed Description
SignedRangeGenotype class.
- Description
- A gene is a double values within the symmetric range specificed in the constructor;
Each gene contained by DoubleGenotype is a double value within the range [-max,+max] where the zero value has two representation: -0 and +0.
The binary is decoded using the first value as sign and the others as absolute value.
- Warnings
Definition at line 39 of file signedrangegenotype.h.
Constructor & Destructor Documentation
| SignedRangeGenotype | ( | unsigned int | numGenes = 0, |
| double | max = +1, |
||
| unsigned int | bitPrecision = 8 |
||
| ) |
Construct a un-initialized SignedRangeGenotype
- Parameters:
-
numGenes is the number of double values contained by this SignedRangeGenotype max is the maximum absolute value represented bitPrecision is the number of bit used for represent a number
- Warning:
- you must explicity call randomize() to get a randomized Genotype
- Note:
- the Genotype::size() return the number of bits of underlying data, while numGenes is the actual numbers of genes of this SignedRangeGenotype. They are related by the multiplication:
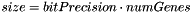
Definition at line 40 of file signedrangegenotype.cpp.
References DoubleGenotype::maxValue().
Referenced by SignedRangeGenotype::clone().
| ~SignedRangeGenotype | ( | ) | [virtual] |
Destructor.
Definition at line 48 of file signedrangegenotype.cpp.
| SignedRangeGenotype | ( | const SignedRangeGenotype & | genotype | ) |
Copy-Constructor (generate a clone of the genoma)
Definition at line 52 of file signedrangegenotype.cpp.
Member Function Documentation
| double at | ( | unsigned int | i | ) | [virtual] |
Return the i-th genes represented.
Implements DoubleGenotype.
Definition at line 123 of file signedrangegenotype.cpp.
References Genotype::bit(), DoubleGenotype::bitprec, Genotype::extractUInt(), and Genotype::size().
| SignedRangeGenotype * clone | ( | ) | const [virtual] |
Create an exact copy of this Genotype.
Reimplemented from Genotype.
Definition at line 152 of file signedrangegenotype.cpp.
References SignedRangeGenotype::SignedRangeGenotype().
| void configure | ( | ConfigurationParameters & | params, |
| QString | prefix | ||
| ) | [virtual] |
Configure the object using the ConfigurationParameters specified.
- Parameters:
-
params the object with configuration parameters prefix the group in which parameters are expected to be (this is terminated by a separator character). We expect to receive as prefix the group with genotype parameters
Reimplemented from Genotype.
Definition at line 68 of file signedrangegenotype.cpp.
References DoubleGenotype::bitprec, Genotype::fitnessv, Genotype::fromCompressedString(), ConfigurationParameters::getValue(), DoubleGenotype::maxv, DoubleGenotype::maxValue(), DoubleGenotype::minv, Genotype::notesv, DoubleGenotype::numgenes, Genotype::rankv, and Genotype::resize().
| void describe | ( | QString | type | ) | [static] |
Add to Factory::typeDescriptions() the descriptions of all parameters and subgroups.
Reimplemented from Genotype.
Definition at line 113 of file signedrangegenotype.cpp.
References ParameterSettable::addTypeDescription(), ParameterSettable::RealDescriptor::def(), ParameterSettable::IntDescriptor::def(), ParameterSettable::Descriptor::describeInt(), ParameterSettable::Descriptor::describeReal(), ParameterSettable::Descriptor::describeString(), ParameterSettable::StringDescriptor::help(), ParameterSettable::RealDescriptor::help(), ParameterSettable::IntDescriptor::help(), ParameterSettable::IsList, ParameterSettable::IsMandatory, ParameterSettable::IntDescriptor::limits(), ParameterSettable::RealDescriptor::props(), and ParameterSettable::IntDescriptor::props().
| SignedRangeGenotype & operator= | ( | const Genotype & | genotype | ) | [virtual] |
Assign operator (performs same operation of Copy-constructor)
Reimplemented from Genotype.
Definition at line 57 of file signedrangegenotype.cpp.
References DoubleGenotype::bitprec, Genotype::copyDataFrom(), DoubleGenotype::maxv, DoubleGenotype::minv, and DoubleGenotype::numgenes.
| void save | ( | ConfigurationParameters & | params, |
| QString | prefix | ||
| ) | [virtual] |
Save the actual status of parameters into the ConfigurationParameters object passed.
- Parameters:
-
params the configuration parameters object on which save actual parameters prefix the prefix to use to access the object configuration parameters.
Reimplemented from Genotype.
Definition at line 98 of file signedrangegenotype.cpp.
References DoubleGenotype::bitprec, ConfigurationParameters::createParameter(), Genotype::fitnessv, DoubleGenotype::maxv, Genotype::notesv, DoubleGenotype::numgenes, Genotype::rankv, and Genotype::toCompressedString().
| void set | ( | unsigned int | i, |
| double | value | ||
| ) | [virtual] |
Set the i-th genes.
Implements DoubleGenotype.
Definition at line 136 of file signedrangegenotype.cpp.
References DoubleGenotype::bitprec, Genotype::insertUInt(), DoubleGenotype::maxv, Genotype::size(), and Genotype::unset().
The documentation for this class was generated from the following files:
- ga/include/genotypes/signedrangegenotype.h
- ga/src/genotypes/signedrangegenotype.cpp